📏K-NN Hyperparamaters#
👨🏫 Vikesh K
📓 Lab 07
💡 “There is no substitute for hard work” 💡
📝Lab Agenda#
We will cover:
sklearn datasets
train-test split with stratification
Model Building with different values of k in k-nn
Visualize the accuracy score for different k
Importing Modules and data#
import numpy as np
import pandas as pd
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from pprint import pprint
import warnings
warnings.filterwarnings("ignore")
print("Modules ran")
Modules ran
💡 We will be using the Wisconsin Cancer Dataset to build a k-nn model
📝 Please read more about the Wisconsin Breast cancer dataset here.[scroll down in the page]
url = 'https://raw.githubusercontent.com/vkoul/data/main/misc/wisconsin.csv'
df = pd.read_csv(url)
df.head()
| diagnosis | radius_mean | texture_mean | perimeter_mean | area_mean | smoothness_mean | compactness_mean | concavity_mean | concave points_mean | symmetry_mean | ... | radius_worst | texture_worst | perimeter_worst | area_worst | smoothness_worst | compactness_worst | concavity_worst | concave points_worst | symmetry_worst | fractal_dimension_worst | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 1 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 1 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 1 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 31 columns
Exploring the data#
df.shape
(569, 31)
df.isna().sum()
diagnosis 0
radius_mean 0
texture_mean 0
perimeter_mean 0
area_mean 0
smoothness_mean 0
compactness_mean 0
concavity_mean 0
concave points_mean 0
symmetry_mean 0
fractal_dimension_mean 0
radius_se 0
texture_se 0
perimeter_se 0
area_se 0
smoothness_se 0
compactness_se 0
concavity_se 0
concave points_se 0
symmetry_se 0
fractal_dimension_se 0
radius_worst 0
texture_worst 0
perimeter_worst 0
area_worst 0
smoothness_worst 0
compactness_worst 0
concavity_worst 0
concave points_worst 0
symmetry_worst 0
fractal_dimension_worst 0
dtype: int64
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 569 entries, 0 to 568
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 diagnosis 569 non-null int64
1 radius_mean 569 non-null float64
2 texture_mean 569 non-null float64
3 perimeter_mean 569 non-null float64
4 area_mean 569 non-null float64
5 smoothness_mean 569 non-null float64
6 compactness_mean 569 non-null float64
7 concavity_mean 569 non-null float64
8 concave points_mean 569 non-null float64
9 symmetry_mean 569 non-null float64
10 fractal_dimension_mean 569 non-null float64
11 radius_se 569 non-null float64
12 texture_se 569 non-null float64
13 perimeter_se 569 non-null float64
14 area_se 569 non-null float64
15 smoothness_se 569 non-null float64
16 compactness_se 569 non-null float64
17 concavity_se 569 non-null float64
18 concave points_se 569 non-null float64
19 symmetry_se 569 non-null float64
20 fractal_dimension_se 569 non-null float64
21 radius_worst 569 non-null float64
22 texture_worst 569 non-null float64
23 perimeter_worst 569 non-null float64
24 area_worst 569 non-null float64
25 smoothness_worst 569 non-null float64
26 compactness_worst 569 non-null float64
27 concavity_worst 569 non-null float64
28 concave points_worst 569 non-null float64
29 symmetry_worst 569 non-null float64
30 fractal_dimension_worst 569 non-null float64
dtypes: float64(30), int64(1)
memory usage: 137.9 KB
Split the dataset into Train and Test
X = df.drop(columns = 'diagnosis')
y = df['diagnosis']
💡Stratified Train-Test Splits
One final consideration is for classification problems only.
Some classification problems do not have a balanced number of examples for each class label. As such, it is desirable to split the dataset into train and test sets in a way that preserves the same proportions of examples in each class as observed in the original dataset.
This is called a stratified train-test split.
We can achieve this by setting the stratify argument to the y component of the original dataset. This will be used by the train_test_split() function to ensure that both the train and test sets have the proportion of examples in each class that is present in the provided “y” array.
Source: MachineLearning Mastery
Train Test Split#
X_train, X_test, y_train, y_test = train_test_split(X, y
,stratify = y # stratification
,random_state = 66 # for reproducibility
,test_size = 0.20)
print(X_train.shape)
print(X_test.shape)
(455, 30)
(114, 30)
Checking the effect of Stratifcation Effect#
Overall Distribution
y.value_counts(normalize = True).round(2).to_frame()
| proportion | |
|---|---|
| diagnosis | |
| 0 | 0.63 |
| 1 | 0.37 |
Train Data Distribution
y_train.value_counts(normalize = True).round(2).to_frame()
| proportion | |
|---|---|
| diagnosis | |
| 0 | 0.63 |
| 1 | 0.37 |
Test Data Distribution
y_test.value_counts(normalize = True).round(2).to_frame()
| proportion | |
|---|---|
| diagnosis | |
| 0 | 0.63 |
| 1 | 0.37 |
👨🏼💻Run the above ratio check after commenting out the stratify argument in train_test_split. Note down your findings
Building the model with 3 neighbours#
You may get errors with the code below if your are using Sklearn version 1.3.0. In that case, you would need to install the version 1.2.2 of sklearn and may also need to install threadpoolctl==3.1.0 to remove the split error
import sklearn
sklearn.__version__
'1.2.2'
# !pip install threadpoolctl==3.1.0
# !pip uninstall scikit-learn
# !pip install scikit-learn==1.2.2
# build the model
clf = KNeighborsClassifier(n_neighbors= 3)
# fit the model
clf.fit(X_train, y_train)
# # record training set accuracy
train_acc = clf.score(X_train, y_train)
print(f"The train data accuracy is {train_acc :.2%}")
# # record test accuracy
test_acc = clf.score(X_test, y_test)
print(f"The test data accuracy is {test_acc :.2%}")
The train data accuracy is 95.82%
The test data accuracy is 91.23%
Accuracy scores across values of k#
We will make use of the for- loop to run k-nn models, varying the n_neighbors argument of the function
# empty list that will be populated
training_accuracy = []
test_accuracy = []
# try n_neighbors from 1 to 51
neighbors_settings = range(1, 51)
for n_neighbors in neighbors_settings:
# build the model
clf = KNeighborsClassifier(n_neighbors= n_neighbors)
clf.fit(X_train, y_train)
# record training set accuracy
training_accuracy.append(clf.score(X_train, y_train))
# record generalization accuracy
test_accuracy.append(clf.score(X_test, y_test))
pprint provides a capability to “pretty-print” arbitrary Python data structures
# pprint(test_accuracy)
# # print("-------")
# pprint(training_accuracy)
len(test_accuracy)
50
len(training_accuracy)
50
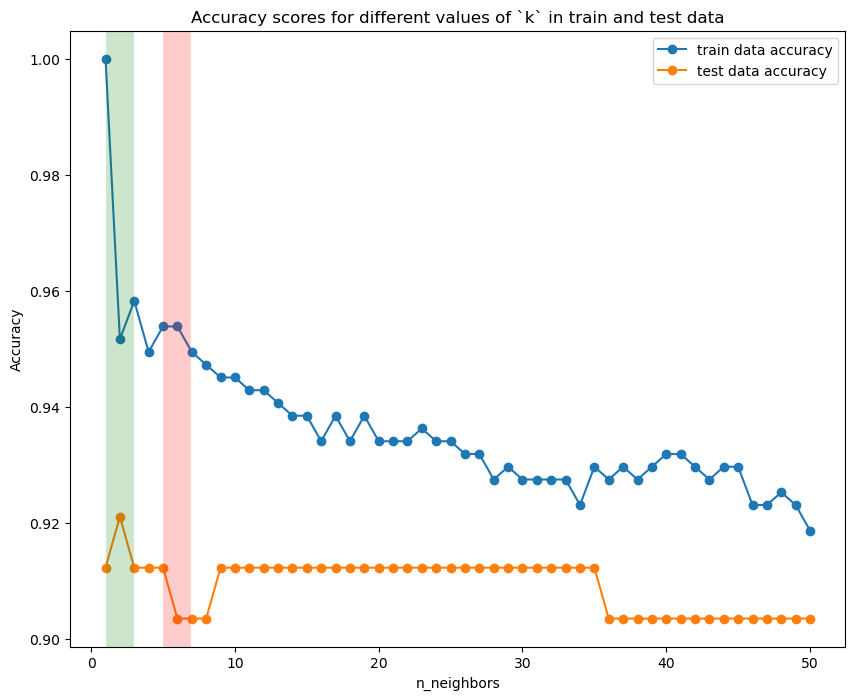
Visualise the results#
plt.figure(figsize= (10, 8))
plt.plot(neighbors_settings, training_accuracy, marker = "o", label = "train data accuracy")
plt.plot(neighbors_settings, test_accuracy, marker = "o", label = "test data accuracy")
# highlight the max and min accuracy values
plt.axvline(x = test_accuracy.index(max(test_accuracy))+1, linewidth = 20, alpha = 0.2, color = "green")
plt.axvline(x = test_accuracy.index(min(test_accuracy))+1, linewidth = 20, alpha = 0.2, color = 'red')
plt.ylabel("Accuracy")
plt.xlabel("n_neighbors")
plt.title("Accuracy scores for different values of `k` in train and test data")
plt.legend();